Lentiviral Particles Production Concentration And Titration
Lentiviral vector harboring the firefly luciferase cDNA under the control of a liver-specific promoter was constructed and used as described earlier. Lentiviral vector particles were packaged with the packaging cassette, NRF, in 293T cells using three-plasmid transient transfection as previously described. Vector titer was determined by measuring the p24 capsid concentration using ELISA as previously described. All vector preps were tested for the absence of replication-competent retrovirus as described earlier.
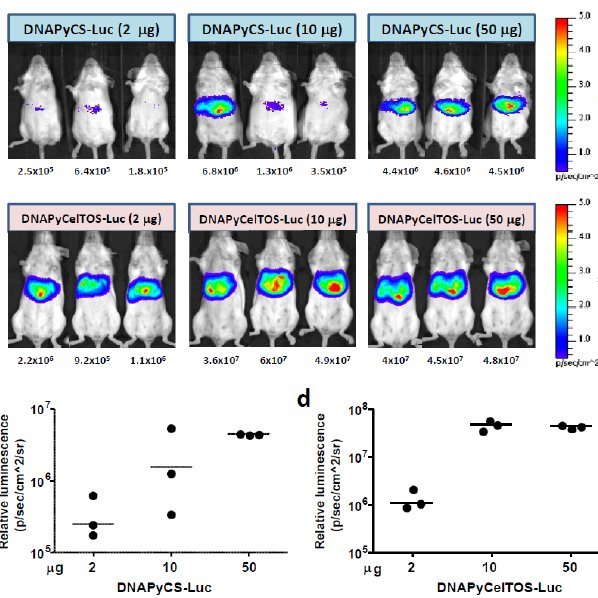
Evaluation Of Luciferase Expression From Cmv Promoter In Vivo
Cytomegalovirus promoters have been widely used to express high levels of exogenous proteins in cultured cells. Similarly, we obtained high expression levels when a plasmid expressing luciferase under a CMV promoter was transfected in HeLa cells . When the same plasmid was introduced in mice by hydrodynamic injection, we also detected very high luciferase activity with a CCD camera . Quantification showed 5 × 1010 luciferase units in the liver of treated mice one day after plasmid administration . However, luciferase activity decreased rapidly and reached a plateau of 5 × 107 luciferase units 20 days after plasmid administration . Thus, there was a 1000-fold decrease in luciferase expression. This reduction in expression has been previously described, and it has been associated with promoter methylation.
Panel A. Plasmid pCMVLuc and p1ATLuc express luciferase from CMV promoter and the 1AT promoter fused to the mouse albumin enhancer, respectively. Panel B. A S/MAR sequence was cloned in the plasmids described in A to generate pCMVLucS/MAR and p1ATLucS/MAR. Amp and Neo denote ampicillin and neomycin resistance genes, respectively.
Panel A. Imaging of light emission. The color gradient is indicated to the right. Panel B. Quantification of the data shown in Panel A. Luciferase units are photons/second × cm2 × sr.
The Luciferase Reporter Assay: How It Works
The luciferase reporter assay is commonly used as a tool to study gene expression at the transcriptional level. It is widely used because it is convenient, relatively inexpensive, and gives quantitative measurements instantaneously.
It has broad applications across various fields of cell and molecular biologywherever you want to measure or track the expression of a cloned gene.
As with many assays and kits, many people who use the luciferase reporter assay dont take the time to fully understand how it works. But understanding the basics of the techniques you are using is essential for troubleshooting when you hit a problem, and for convincing your thesis examiners that you know what you are talking about!
So lets dive into the inner workings of the luciferase assay.
Don’t Miss: Hepatitis B And Rheumatoid Arthritis Treatment
Mortality And Atypical Pathological Responses Across This Treated Cohort
A total of 12/61 mice within this study died during the course of this experiment. This mortality impacted seven strains , with CC021/Unc experiencing the highest mortality . After sacrifice, six mice from four strains showed what appeared to be a macroscopic abnormality, including liver and spleen tumors. Pathological analysis suggested these were common lesion types in aging animals and unlikely to be due to the presence of vector-related issues .
Tg And Tch Analysis And Alt/ast Activity Assay
Alanine aminotransferases assay kit, aspartate aminotransferases assay kit, triglyceride and total cholesterol assay kits were from Jiancheng Biology Institution PeproTech . The serum or liver tissue levels of TG and TCH in mice were assayed using TG and TCH assay kits according to the protocols. ALT and AST levels in serum from C57BL/6J mice with ALD were analyzed by using ALT and AST activity assay kits according to the protocols recommended by the manufacturer. The absorbance was measured at 510 nm.
Also Check: What Are The Symptoms To Hepatitis C
Bioinformatics Analysis And Chromatin Immunoprecipitation
We used regulatory sequence analysis tools to analyze the sequence of the SREBP2 gene promoter to find high-score ERE-like sequences. HepG2 and HuH-7 cells were treated with 107mol/l E2 for 24h and then cross-linked according to the Millipore EZ-ChIP Assay Kit protocol .
Immunoprecipitation was performed with the following antibodies purchased from Millipore: mouse anti-human ER ChIP antibody, mouse IgG used as the negative control, and mouse anti-human RNA Polymerase II antibody used as the positive control.
SREBP2 was then detected via PCR using 5-GTCTCCAACTCCTGACCTCAA-3 and 5-AGTGCCTTGCATACTGCTGTA-3 as the primer sequences. The PCR products were analyzed using agarose electrophoresis and the band was excised from the gel. Finally, the PCR product was sequenced by Invitrogen.
Relationship Between Vcn And Luciferase Expression
Given the highly significant variation in total and liver-specific luciferase expression between this collection of mouse strains, we sought to assess the extent to which host genetic differences controlled these expression levels . We determined the relative expression of luciferase at week 41 within the livers of each mouse relative to liver VCN. We found that vector-specific activity in mouse livers was impacted by the host strain . Heritability analysis showed that between 15% and 27% of the variation in vector-specific activity in the liver was explained by host genetic variation. We utilized a second analysis to assess the impact of host genetic variation on luciferase expression independent of host genetic variations impact on vector copy number. We used a partial F-test to assess the improvement in fit when our model explaining luciferase levels included both strain and vector copy number as opposed to vector copy number alone. We found strong evidence for liver luciferase expression having other host factors driving expression , although the evidence for total luciferase expression was not significant .
Read Also: What Is Chronic Viral Hepatitis C
Multiple Regulatory Mechanisms Exist For Hepatic Dec1 Expression
First we determined the effects of Clock mutation on hepatic expression of and . In wild-type mice , hepatic expression of mRNA showed a circadian rhythm with peaks at ZT 6 , whereas in Clock mutant mice , peaks were delayed by 6 h compared with the peak in wild-type mice, but the pattern maintained a significant rhythmicity . Differences in the patterns between genotypes were significant indicating that CLOCK is indeed involved in the regulation of but there seem to be some other mechanism . On the other hand, strong circadian rhythm of expression with a peak at ZT6 was observed in wild-type mouse liver , and this was abolished by Clock mutation , indicating that CLOCK : BMAL1 heterodimer dominantly regulates expression .
Figure 1
Lxr and Lxr expression in the liver, kidney, and heart of mice. Relative levels of Lxr and Lxr mRNA were determined by quantitative real-time RT-PCR . Data were plotted as mean ± SEM . , Western blot of LXR protein in the nuclear extracts of the three tissues. A representative photograph is shown and quantitative values were plotted as mean ± SEM . The size of LXR protein is indicated.
Efficient Hepatic Delivery And Protein Expression Enabled By Optimized Mrna And Ionizable Lipid Nanoparticle
mRNA therapy represents a promising revolutionary treatment strategy.
-
Sequence optimization and efficient delivery are critical to its wide application.
-
Ionizable lipid nanoparticle was developed to deliver codon-optimized mRNA.
-
Robust protein expressions of luciferase and erythropoietin were achieved both in vitro and in vivo.
-
ILP171/mRNA exhibited excellent safety profiles in vivo.
Don’t Miss: Where Do You Get Hepatitis C
Firefly And Click Beetle
The luciferases of fireflies of which there are over 2000 species and of the other Elateroidea are diverse enough to be useful in molecular phylogeny. In fireflies, the oxygen required is supplied through a tube in the abdomen called the abdominaltrachea. One well-studied luciferase is that of the Photinini firefly Photinus pyralis, which has an optimum pH of 7.8.
How Does Luciferase Reporter Assay Work
To perform a luciferase reporter assay, you will need a DNA plasmid to express the protein that you hypothesize could affect transcription.
You will also need to use another DNA plasmid with the regulatory element fused with the DNA coding sequence for a luciferase enzyme. When activated, this system produces luciferase.
After cells are transfected with the luciferase reporter plasmid andallowed to grow for few days, the next steps are to lyse the cells, add asubstrate to the cell lysate, and measure the luciferase activity based on theamount of bioluminescent signal.
A Luciferase Reporter Assay. The luciferase assay is useful to study whether a protein of interest regulates a particular gene at the transcription level. By transfection, a DNA construct with the genes promoter and a coding region of the luciferase reporter gene enters the cells. Another DNA construct introduced into the cells consists of a coding region of the protein of interest. When this protein activates transcription, the cell will produce luciferase enzyme. After the addition of a lysis buffer and a substrate, a luminometer quantifies the luciferase activity.
If your protein activates the expression of the target gene, the amount of signal produced increases. However, if it blocks the gene expression, the cells produce less luciferase. As a result, these samples generate a lower signal than the positive control.
You May Like: Royal Canin Hepatic Wet Dog Food
Examples Of Luciferase In A Sentence
luciferase Quanta Magazineluciferase USA TODAYluciferase Scientific AmericanluciferaseEssenceluciferaseEssenceluciferase baltimoresun.comluciferase Travel + LeisureluciferaseUSA TODAY
These example sentences are selected automatically from various online news sources to reflect current usage of the word ‘luciferase.’ Views expressed in the examples do not represent the opinion of Merriam-Webster or its editors. Send us feedback.
What Information Does A Luciferase Assay Tell You

The luciferase reporter assay allows you to study the regulatory control of a gene of interest. The presence of light means that luciferase was transcribed, and therefore the regulatory region was active. This can allow you to determine if a particular protein is involved in the expression of your gene of interest or if the regulatory region of your gene of interest is active under certain conditions.
You May Like: Hepatitis B Is More Infectious Than Hiv
Intersection Between Yy1 Chip
The presence of YY1 binding nearby the DEGs can be indicative of a direct regulatory role of YY1 in regulating the DEGs expression. To achieve better understanding of the molecular mechanisms regulating the transcriptional alternations induced by YY1 knockdown, we obtained YY1 chromatin immunoprecipitation coupled with massively parallel sequencing data in HepG2 cells and liver tissues from the ENCODE project . In accordance with previous observations that YY1 commonly occupied active promoters and enhancers, the reads in YY1 ChIP-seq experiments from both HepG2 cells and liver tissues were highly enriched in the promoter regions . By intersecting the DEGs with their nearby YY1-binding peaks defined in HepG2 cells, we identified 596 down-regulated genes bearing 902 YY1 peaks nearby and 620 up-regulated genes bearing 933 YY1 peaks nearby . We further divided the DEGs based on their overall deregulation and the presence of YY1 peaks. shows that of the DEGs that have YY1 bound nearby, which are presumed to be the direct targets of YY1, approximately equal proportions of the up-regulated and down-regulated genes are evenly distributed based on their changes in expression level. A larger fraction of the up-regulated genes did not have YY1 bound nearby, especially for the DEGs that are up-regulated more than fourfold by YY1 knockdown.
Mouse Tissue Dna Preparation For Quantification Of Vector Copy Number
At the end of the experiment, the viral copy number of liver and spleen tissues was determined. Genomic DNAs from tissues were isolated using the Blood & Tissue DNeasy kit according to the manufacturers instructions. RNAs were removed using RNase A . All samples were treated with DpnI . Total vector copy number of each sample was quantified using multiplex qPCR, as previously described.
Read Also: Pro Plan Hepatic Dog Food
Loss Of Hepatic E4bp4 Protects Against Hflmcd Diet
Given the important role of ER stress in hepatocytes during NAFLD and the elevated E4BP4 expression in the liver of the HFLMCD diet-induced ER stress mouse model , it was logical to examine whether manipulation of hepatic E4BP4 impacts lipid metabolism and NAFLD in vivo. We first compared E4bp4flox/flox vs E4bp4-LKO mice on regular chow following fasting overnight and refeeding for 12 hours. The reason we chose to refeed mice is because the E4BP4 mRNA and protein tend to be potently induced by refeeding. Body weight, TG, and cholesterol in serum/liver, as well as serum ALT all turned out to be comparable between the two groups of mice , supporting that hepatic E4BP4 is dispensable in liver lipid metabolism under the normal physiological condition.
Next, we examined the impact of hepatic deletion of E4bp4 on lipid metabolism following HFLMCD diet. Upon 10-wk HFLMCD feeding, both E4bp4flox/flox and E4bp4-LKO mice showed similar body weight gain . However, the ratio of liver weight/body weight was significantly reduced in E4bp4-LKO mice . Serum ALT, the classical marker for liver injury, was about 20% lower in E4bp4-LKO mice . Despite comparable levels of serum TG and cholesterol in both groups of mice , E4bp4-LKO mice accumulated significantly less TG, total cholesterol, and lipid droplets in the liver by H& E staining . In summary, all these data suggest that E4bp4-LKO mice are protected from HFLMCD-induced liver steatosis and cholesterol accumulation.
FIGURE 5E4bp4PP
Reverse Transcription And Quantitative Real
Two micrograms of total RNA was reverse transcribed using 10 M each of dCTP, dGTP, dATP, and dTTP, 100 ng random primers, 1 × first-strand buffer , 0.01 M DTT, and 200 units of SuperScript II reverse transcriptase in a 20 L reaction for 90 minutes at 42°C. Real-time polymerase chain reaction was performed using the ABI Prism 7500 Applied Biosystems . Amplification reactions were carried out in 25 L volume using SYBR Green I dye and the following amplification conditions: 50°C for 2 minutes and 95°C for 10 minutes for 45 cycles. Primers were designed to specifically amplify 123 bp of hepcidin cDNA 113 bp of GAPDH cDNA 152 bp of STAT3 cDNA and 127 bp of C/EBP cDNA .
The mRNA/cDNA abundance of each gene was calculated relative to the expression of a housekeeping gene, GAPDH .
Recommended Reading: New Drug To Cure Hepatitis C
Immunization Of Mice With Radiation Attenuated Sporozoites Or Sporozoites Under Chloroquine Prophylaxis
C57BL/6 mice were immunized with wildtype P. berghei radiation attenuated sporozoites or sporozoites under chloroquine prophylaxis . Immunizations were performed by i.v injection with three doses of 1 × 104 or 4 × 103 sporozoites, with a 7 day interval between the boosts. For CPS immunization, mice received 800 g chloroquine base in PBS i.p, starting from sporozoite injection up to two weeks after the last immunization. Absence of blood stage parasites was confirmed by examination of Giemsa-stained blood smears of tail blood at the end of the chloroquine treatment period and approximately 1 day before challenge. Mice were challenged two weeks after ending choroquine treatment. Irradiation of sporozoites was performed by exposure of infected A. stephensi mosquitoes to 16,000 rad of -radiation .
Hdac4 Regulates Fibrogenesis By Targeting Mir
Since HDAC4 silencing blocked the induction of MRTF-A protein levels by TGF- and PDGF , we asked whether HDAC4 might regulate fibrogenesis by targeting miR-206. Over-expression of HDAC4 activated the promoter activities of key pro-fibrogenic genes, including Col1a1 and Col1a2, in HSC-T6 cells . This effect, however, was essentially neutralized by the introduction of miR-206 mimic and restored by MRTF-A over-expression . On the other hand, qPCR analyses demonstrated that while HDAC4 knockdown attenuated the induction of endogenous levels of collagen type I and type III by TGF- and PDGF, miR-206 antimir reversed this trend and restored the expression of pro-fibrogenic genes . Therefore, we propose that HDAC4-mediated suppression of miR-206 may play a role in liver fibrogenesis.
You May Like: What Is A Hepatitis Panel
Protein Isolation And Western Blot Analysis
Protein was extracted from AML-12 cells or liver tissues using RIPA lysis buffer supplemented with 1% PMSF. After centrifugation for 30 min at 4°C at 12000 rpm, upper supernatant was collected. Protein was separated on 8â12% SDS-polyacrylamide mini-gels, then transferred onto polyvinylidene fluoride membranes . After blocked in 5% non-fat milk for 3 h, membranes were washed in TBS-Tween20 buffer three times per 10 min, and incubated with specific primary antibodies overnight. Primary antibodies Lipin1 , PPAR-α , SREBP-1 , FASN were purchased from Abcam. Primary antibodies IL-6 was purchased from Bioworld. β-Actin and TNF-α were purchased from Bioss. Histone3 was purchased from Proteintech. Then the membranes were washed for three times with TBS-Tween20 buffer, following incubation with the corresponding horseradish peroxidase conjugated secondary antibody for 1 h and then developed onto chemiluminescence. Western blotting detection system using ECL-chemiluminescent kit . Quantitative densitometric analyses of immunoblotting images were performed using ImageJ software. The experiment was repeated for three times.
The Lncrna Gm15622 Stimulates Srebp

- 1 M. Ma and R. Duan contributed equally to this work.Minjuan Ma1 M. Ma and R. Duan contributed equally to this work.AffiliationsJiangsu Key Laboratory for Molecular and Medical Biotechnology, School of Life Science, Nanjing Normal University, Nanjing 210023, China
- 1 M. Ma and R. Duan contributed equally to this work.Rui Duan1 M. Ma and R. Duan contributed equally to this work.AffiliationsJiangsu Key Laboratory for Molecular and Medical Biotechnology, School of Life Science, Nanjing Normal University, Nanjing 210023, China
- Lulu ShenAffiliationsJiangsu Key Laboratory for Molecular and Medical Biotechnology, School of Life Science, Nanjing Normal University, Nanjing 210023, China
- Mengting LiuAffiliationsJiangsu Key Laboratory for Molecular and Medical Biotechnology, School of Life Science, Nanjing Normal University, Nanjing 210023, China
- Yaya JiAffiliationsJiangsu Key Laboratory for Molecular and Medical Biotechnology, School of Life Science, Nanjing Normal University, Nanjing 210023, China
- To whom correspondence should be addressed.
Cell. Mol. Life Sci.
Nat. Rev. Gastroenterol. Hepatol.
Read Also: How Do You Test For Hepatitis C