Why Was This Study Done
An effective way to limit the global spread of HCV might be to introduce routine screening of the blood that is used for transfusions in developing countries. In developed countries, HCV screening of blood donors use expensive, commercial RT-PCR assays to detect small amounts of HCV ribonucleic acid . All the current HCV assays, which can also quantify the amount of viral RNA in the blood during treatment, detect a target sequence in the viral genome called the 5-noncoding region . However, there are several different HCV genotypes . These genotypes vary in their geographical distribution and, even though the 5-NCR sequence is very similar in the common genotypes , the existing assays do not detect all the variants equally well. This shortcoming, together with their high cost, means that 5-NCR RT-PCR assays are not ideal for use in many developing countries. In this study, the researchers identify an alternative diagnostic target sequence in the HCV genomethe 3-X-tail elementand ask whether this sequence can be used to develop a new generation of tests for HCV infection that might be more appropriate for use in developing countries.
Time For Processing Hcv Ab Test Results
The turnaround time for 3rd-generation EIAs is at least 1 day. Many labs do not perform the tests on site and must send specimens to another lab for processing, which may further increase the turnaround time.
A point-of-care test is also available. The OraQuick® HCV Rapid Antibody Test is an FDA-approved test that can be performed with a fingerstick . It is also a CLIA-waived test and therefore can be used in clinic offices and outreach facilities. Results are reported as reactive or nonreactive within 20 minutes. Just as for the standard HCV Ab test done in the lab, a positive OraQuick® test must be confirmed by an HCV RNA test. The sensitivity and specificity of the test is similar to that of the laboratory-based assays.
Evaluation Of A Reverse Transcriptase Loop Mediated Isothermal Amplification Assay For Detection Hepatitis C Genotypes 1
Goreti Atieno Omolo,1 Hans Nitschko,3 Raphael Lwembe,2 Alex Kattam Maiyo,1,2 Eddy Okoth Odari1
Verify Captcha
Regret for the inconvenience: we are taking measures to prevent fraudulent form submissions by extractors and page crawlers. Please type the correct Captcha word to see email ID.
0#Both authors have equally contributed to this study1Department of Medical Microbiology, Jomo Kenyatta University of Agriculture and Technology, Kenya2Center for Virus Research, Kenya Medical Research Institute, Kenya3Diagnostic virology department, Max von Pettenkofer-Institute , Germany
Correspondence: Eddy Okoth Odari, Department of Medical Microbiology, Jomo Kenyatta University of Agriculture and Technology P.O. Box 62000-00200. Nairobi, Kenya
Citation: Omolo GA, Nitschko H, Lwembe R, Maiyo AK, Odari EO Evaluation of A Reverse Transcriptase Loop Mediated Isothermal Amplification Assay For Detection Hepatitis C Genotypes 1-4 Viruses Under Limited Logistical Conditions. J Hum Virol Retrovirol 5: 00145. DOI: 10.15406/jhvrv.2017.05.00145
Read Also: Interpretation Of Hepatitis A Serologic Test Results
Optimal Concentration Of Ic
Armored RNA was serially diluted and then spiked into the national reference material for HCV RNA . Armored RNA was coextracted and coamplified with the samples in the same reaction tube. According to the results presented in Table , 1000 copies/ml of armored RNA was used as the optimal concentration of IC in the HCV RNA duplex real-time RT-PCR assay.
Table 1 Optimization of the concentration of IC
What Do These Findings Mean

These findings suggest that the HCV 3-X-tail element could provide an alternative target for screening blood samples for HCV infection and for monitoring viral loads during treatment, irrespective of HCV genotype. In addition, they suggest that X-tail RT-PCR assays may be stable and robust enough for use in laboratories in emerging countries. Overall, these findings should stimulate the development of a new generation of clinical HCV assays that, because the protocol used in the X-tail assay is freely available, could improve blood safety in developing countries by providing a cheap and effective alternative to existing proprietary HCV assays.
Don’t Miss: Can Alcohol Cause Hepatitis C
Nucleic Acid Amplification Technologies
2.1. Non-Isothermal Amplification Methods
2.1.1. Reverse Transcription Polymerase Chain Reaction
2.1.2. Real-Time PCR
2.2. Isothermal Amplification Methods
2.2.1. Nucleic Acid Sequence-Based Amplification
2.2.2. Transcription-Mediated Amplification
2.2.3. Reverse Transcription Loop-Mediated Isothermal Amplification
2.2.4. Rolling Circle Amplification Method
Overview Of The Hepatitis B Virusloop
The principle of HBV-LAMP-LFB assay is illustrated in Figure 1. In brief, the HBV genomic DNA was extracted using nucleic acidreleasing agents within 10 min, and then, preamplification was carried out at a fixed temperature for 40 min. We modified the LAMP primers LF and FIP at 5-end with biotin and FAM, respectively. The HBV-LAMP amplicons were labeled with biotin and FAM finally, the LAMPs were read out with LFB . For biosensor detection, the dye streptavidin-coated gold nanoparticles were laminated onto the conjugate pad, by adding 1.0 L of HBV-LAMP products and 100 µL of running buffer to the sample pad. The running buffer containing HBV-LAMP products were absorbed, and the detection results were read out visually on the NC membrane within 2 min. In the positive sample, the biotin/FAM-labeled HBV-LAMP amplicons were captured by anti-FAM at the TL and streptavidin-GNPs were captured by biotin-BSA at the CL. However, in negative results, only the streptavidin-GNPs were captured the biotin-BSA at the CL .
You May Like: How Can I Get Hepatitis
Detection Of Antimicrobial Resistance
As many of the genetic mechanisms of antimicrobial resistance have become better understood, nucleic acid amplification methods have proved to be useful for the confirmation of antimicrobial resistance in laboratory isolates and for the direct detection of such resistance in clinical specimens. Conventional culture and susceptibility test procedures for most pathogenic bacteria generally take 48-72 hours. The performance of these tests may be erratic because factors such as inoculum size or variability in culture conditions may affect phenotypic expression of resistance. Amplification of genetic determinants may therefore be used to confirm antimicrobial resistance based on the organism’s genotype rather than relying on the variability of phenotypic expression of the resistance . Moreover, these tests can be done within hours, providing clinically relevant information days before conventional susceptibility test results become available. Molecular assays to detect antimicrobial resistance directly from clinical samples have also been described.,
Table 3.
Screening For Hcv Infection
HCV screening has several potential benefits. By detecting HCV infection early, antiviral treatment can be offered earlier in the course of the disease which is more effective than starting at a later stage. Further, early detection together with counseling and lifestyle modifications may reduce the risk of transmission of HCV infection to other people. The optimal approach to screen for HCV is to test the individuals having risk factors for exposure to the virus. The American Association for the Study of Liver Diseases recommends screening for HCV for the following individuals:
-
Recipient of blood or blood components .
-
Recipient of blood from a HCV-positive donor.
-
Injection drug user .
-
Persons with following associated conditions
-
persons with HIV infection,
-
persons who have ever been on hemodialysis, and
-
persons with unexplained abnormal aminotransferase levels.
-
Children born to HCV-infected mothers.
-
Healthcare workers after a needle stick injury or mucosal exposure to HCV-positive blood.
-
Current sexual partners of HCV-infected persons.
Also Check: How Does One Get Hepatitis B And C
Feasibility Studies And First Results
Initial publications on the application of in-house approaches for mini pool plasma testing confirmed concerns about the maturity of PCR testing in a blood bank setting. PCR was well established for patient diagnostics however, screening of thousands of healthy and mostly negative blood donors per day required quite different approaches. Repeat testing was no option and therefore false negatives had to be avoided. Thus, screening of blood donors required the highest sensitivity and specificity, highest throughput, shortest time to result, and lowest costs, which altogether were quite challenging.
The usefulness of the direct virus detection by PCR for blood donor screening was already reported in the mid-1990s for single donation testing . In 1998, the first data were published on the routine screening of mini pools of up to 600 donations for HCV, HBV, and HIV-1 by PCR, with a throughput of 3,000 samples per day. Among 430 thousand donations tested and negative in serological assays, 0 were HIV-1, 2 were HBV, and 22 were HCV positive by PCR only . These high numbers of HCV PCR-only positives exceeded by a factor of 10 the estimated yield rate of HCV seronegative/PCR positive donations in Germany according to previous publications . However, a further study performed on smaller pools of 96 samples reported no viremic seronegative donation among 332 thousand tested, which was below the expected number .
The Standard Hepatitis B Virusloop
The HBV-LAMP was carried out in a 25 L reaction system, including 12.5 L of 2 × reaction buffer , 40 mM KCl, 16 mM MgSO4, 20 mM 2SO4, 2 M betaine, and 0.2% Tween-20), 1 L genomic template, 0.4 M each of outer primer F3 and B3, 1.6 M each of inner primer FIP* and BIP, 0.8 M each of loop primer LF* and LB, 1 L of Bst 2.0 DNA polymerase, 1 L of AMV reverse transcriptase , and 1 µl colorimetric indicator , along with addition of double distilled water to 25 L. The LAMP was performed at 64°C for 1 h. HCV and HIV were used as negative controls , and double distilled water was used as the template in blank control .
Read Also: If You Have Hepatitis B Do You Have Hiv
What Did The Researchers Do And Find
The researchers determined the RNA sequence of the 3-X-tail element in reference samples of the major HCV genotypes and showed that this region of the HCV genome is as highly conserved as the 5-NCR. They then developed a prototype X-tail RT-PCR assay and tested its ability to detect small amounts of HCV and to measure viral load in genotype reference samples and in a large panel of HCV-infected blood samples collected in Germany, the UK, Brazil, South Africa, and Singapore. The new assay detected low levels of HCV RNA in all of the genotype reference samples and was also able to quantify high RNA concentrations. The viral load estimates it provided for the clinical samples agreed well with those obtained using a commercial assay irrespective of the sample’s HCV genotype. Finally, the X-tail RT-PCR assay gave similar results to a standard assay at a fraction of the cost when used to measure viral loads in a Brazilian laboratory in an independent group of 127 patient samples collected in Brazil.
Terms And Abbreviations Used In This Report
ALT Additional testing automatically performed in response to a screening-test–positive result RIBA® Recombinant immunoblot assay, a more specific serological anti-HCV assay RT-PCR Reverse transcriptase-polymerase chain reaction amplification, a nucleic acid testing method for detection of HCV RNA S/Co ratio Signal to cut-off ratio, calculated by dividing the OD value of the sample being tested by the OD value of the assay cut-off for that run Screening tests Serologic immunoassays for detection of anti-HCV Screening-test–negative
Screening immunoassay test result interpreted as negative on the basis of criteria provided by the manufacturer
Screening-test–positive Screening immunoassay test result interpreted as positive on the basis of criteria provided by the manufacturer STD Sexually transmitted disease Supplemental test More specific test used to verify a positive anti-HCV screening test result TMA
You May Like: Hepatitis B Surface Antibody Positive
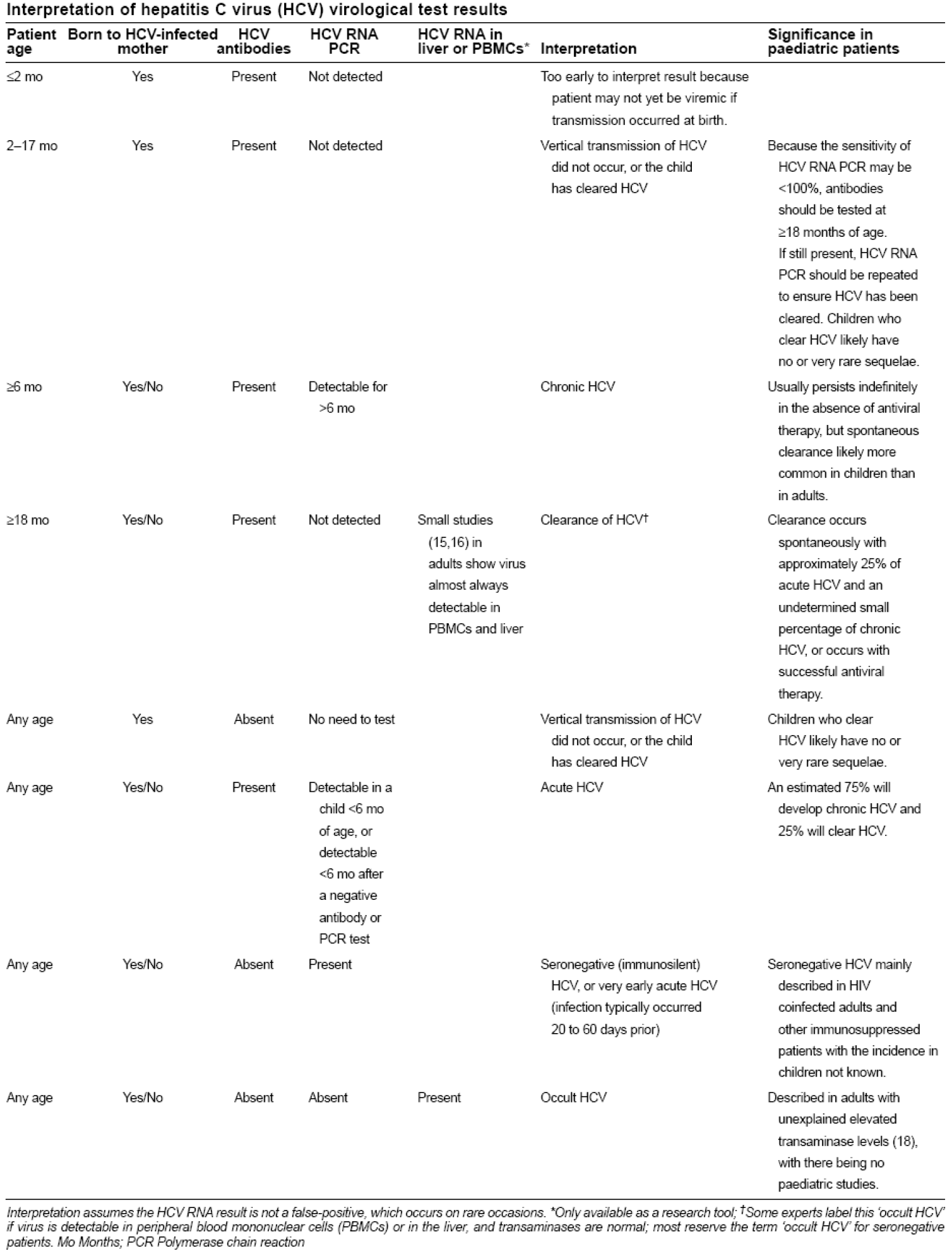
Interpreting Hcv Rna Test Results
It is essential that the provider understands how to interpret HCV RNA test results, especially during the course of HCV treatment.
| Result of HCV RNA Test | Interpretation |
|---|---|
| A quantified viral load — any exact number | Ongoing HCV infection |
| “Detected” | The HCV RNA is detectable but the number of international units is so low that it cannot be quantified accurately. This indicates extremely low level of virus is present. |
| “< 12 IU/mL” or “< 15 IU/mL” or “< 25 IU/mL” All of these are “less than the LLOQ” | HCV RNA is undetectable. No virus is detected at all in the patient’s serum specimen. |
Fluorescence Detection And Spectrofluorometric Analysis Of Products
In order to investigate as to whether the virus-specific fluorogenic oligonucleotides hybridized to their targets, fluorescence intensities in reaction tubes were evaluated with the naked-eye using a handheld UV-transilluminator at 302nm. Reaction tubes were photographed using a BlackBerry Z30 smartphone camera and the iPad Air tablet camera . Additionally, fluorimetric evaluation of fluorescence intensity was performed by measuring 1.5L of target and control amplification products on a NanoDrop-3300 spectrofluorometer .
Recommended Reading: Does Hepatitis C Have A Vaccine
Construction Of The Gold Nanoparticle
The gold nanoparticle-based LFB used in the current study is devised and elaborated in Figure 2. In brief, the biosensor consists of four sections, including sample pad, conjugate pad, nitrocellulose membrane, and absorbent pad . The detector reagents , 129 nm, 10 mg·ml1 100 mM borate, pH 8.5 with 0.05% Tween 20 0.1% BSA and 10 mM EDTA) were laminated onto the conjugate pad. Biotin-BSA and rabbit anti-FAM were coupled onto the NC membrane for the control line and test line , respectively. Each band was separated by 5 mm. In the end, the assembled biosensor was preserved in a plastic box and conserved with a silica gel desiccant at room temperature.
Incorporation Of Dna Amplification Technology Into The Diagnostic Microbiology Laboratory
Despite the obvious advantages to these newer procedures, there may be potential limitations to DNA amplification technology in the diagnostic microbiology laboratory . The accuracy and reproducibility of PCR assays depend on the technical expertise and experience of the operator. Specificity of the test may be affected by contamination of the specimen during laboratory processing, if nonspecific primers are selected for the assay or if PCR conditions are not optimal, allowing nonspecific products to amplify. The most common sources of contamination are from other samples or from previous amplification procedures. Contamination or amplification product carry-over of even minute amounts of nucleic acid may result in the generation of billions of DNA copies that may lead to a false-positive test result. For this reason laboratories should have separate rooms for different steps of the PCR procedure and must follow stringent quality control measures to prevent contamination or carry-over. False-negative test results may occur because of the presence of substances in the specimen that inhibit nucleic acid extraction or amplification. Certain specimen types are more likely to contain such inhibitors. The assays may also lack sensitivity if there is a low inoculum of the microorganism present in the clinical specimen. This may be exacerbated if an inadequate sample or very small specimen volume is available for testing.
Table 4.
You May Like: Hepatic Vein Thrombosis Treatment Guidelines
Risk Of Hcv Infection In Recipients Of Blood Transfusion
Prior to 1992, blood transfusions carried a high risk of HCV infection, approximately 15-20% with each unit transfused. In 1988, 90% of cases of posttransfusion hepatitis were due to NANBH viruses which was later found out to be due to HCV. The move to all-volunteer blood donors instead of paid donors had significantly reduced the risk of posttransfusion hepatitis to 10%. Screening of blood further reduced the rate of posttransfusion hepatitis C by a factor of about 10,000 to a current rate of 1 per million transfusions. The few cases that still occur are due to newly infected people donating blood before they have developed antibodies to the virus, which can take up to 6-8 weeks.
Amplified Nucleic Acid Techniques
The use of molecular diagnostic methods dates back to 1980s via progression in PCR. Accuracy and rapidity are the most important aims in research and clinical diagnostics. According to different studies, there are several methodologies in the field of nucleic acid amplification, which are based on probe, signal, or target .
7.2.1. Probe Amplification Techniques
In this category of the hybridization consisting of probe and target nucleic acid sequence, several copies are constructed. The isothermal nature of probe amplification techniques is their main advantage. It is important to know that each probe amplification technology has its particular properties therefore, each technology is applied for a particular sample diagnostics. There are different probe amplification methods. The most common techniques for hepatitis viruses detection are cycling probe technology , invader assay, and ligase chain reaction .
7.2.1.1. Cycling Probe Technology
CPT is an isothermal signal amplification technique that is proper for detecting low amounts of DNA as a target sequence. The principle of CPT is constructed on the activity of a thermostable enzyme of RNase H and the presence of a chimeric probe of DNA-RNA-DNA with 25- to 30-kb length. CPT is a simple and linear DNA detection technique .
7.2.1.1.1. Advantages and Disadvantages
7.2.1.2. Invader Assay
7.2.1.2.1. Advantages and Disadvantages
7.2.1.3. Ligase Chain Reaction
7.2.1.3.1. Advantages and Disadvantages
7.2.2.1. PCR Techniques
Also Check: Hepatitis B What You Need To Know
Looking Further: Hcv Vaccines
Vaccine development for HCV is currently one of the most challenging fields in virology today. Various obstacles that hinder the development of an effective preventive or therapeutic vaccine for HCV include:
Considerable genetic heterogeneity of isolates within and between geographic locales .
Evolution and existence of quasispecies in an individual .
Poorly defined immunological correlates of protection.
Lack of efficient in vitro propagation to isolate the virus.
Despite these obstacles, both preventive as well as therapeutic vaccines for HCV are under development and also under various phases of vaccine trials, but a successful vaccine remains to be developed.
Confirming The Feasibility Of The Hepatitis B Virusloop

In order to confirm the applicability of HBV-LAMP-LFB assay in clinical setting, 115 serum samples with suspected HBV infection were collected from the Second Affiliated Hospital of Guizhou University of Traditional Chinese Medicine. All of the clinical specimens were evaluated simultaneously using qPCR and HBV-LAMP-LFB. The qPCR diagnosis was carried out using a commercial real-time TaqMan PCR kit , and the detection was carried out with the Applied Biosystems 7,500 Real-Time PCR system . The concentrations of HBV less than 30 IU will be considered as the negative result according to the manufacturers instructions. The HBV-LAMP-LFB operation is as described above.
In the current study, each test was repeated at least three times for repeatability.
You May Like: How Can You Contact Hepatitis C